The DNA testing service matches people with identical or similar DNA. I am in communication with a Mr. Robert Leblanc who has an identical "H" Haplogroup. We are comparing notes about our genetic history.
We have also been matched with a number of other individuals with similar mtDNA. Over time we will compare notes to determine our relationship.
I will post our detailed genealogy GedCom file when I get back home which will help facilitate connections. Beth and I a traveling now - see our recent trips at:
__________________
WHAT DOES MY H SUBCLADE MEAN?
Three recent papers published in Molecular Biology and Evolution, The American Journal of Human Genetics, and Genome Research and led by Dr. Eva-Liis Loogväli and Professor Richard Villems in Tartu, Estonia, and Dr. Alessandro Achilli and Professor Antonio Torroni in Pavia, Italy, and Dr. Luisa Pereira in Porto, Portugal, respectively, represent an advance in understanding in haplogroup H, which represents about 40% of all maternal lineages in Europe, and stretches into Western Asia as well.Because this haplogroup is dominant in West Eurasia, Family Tree DNA began offering testing for sub-haplogroups H1-11 and downstream variants in conjunction with the first published paper, entitled Disuniting Uniformity: A Pied Cladistic Canvas of mtDNA haplogroup H in Eurasia. We expect that this first anthropological attempt to offer genuine resolution for H-clade mitochondria will begin to answer the question that has been asked time and again on our female lineages: where (even approximately) do we come from; or because Europe was re-settled after the end of the last Glacial Maximum, more appropriately, where is my mtDNA most concentrated in Eurasia.
The paper defines Europeans as falling into 10 haplogroups, known as H, J, K, N1, T, U4, U5, V, X and W. Of interest also was the finding that base pairs 16093 and 16311 were the most variable being found in 7 different sub-clades of haplogroup H. We now extend our genotyping to H1-H15 based on information drawn from these papers mentioned above.
The papers use both control (aka HVR-1 and HVR-2) and coding region mutations to organize the phylogeny of haplogroup H and offer some interesting geographical information not discussed in print before. Further the papers highlight the most polymorphic sites with clade H, based on fast, average and slow mutating sites from within the HVR control region. It’s clear from the study that the wave of the future will be full genome scanning for mitochondria to offer the type of resolution that we have become accustomed to when looking at the non-recombining Y chromosome.
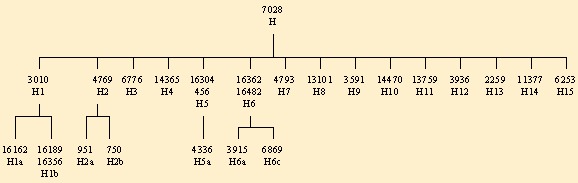
The chart at the top of this page represents a combination of the results from these papers. The second paper, regarding Torroni's work, made minor corrections to the arrangements of the sub-clades on the H sub-clade tree; the chart on this page represents the corrected version.
One interesting note in the papers was that the ancestral haplogroups of H, R0 (formerly Pre-HV) and HV*, appear to be Near/Middle Eastern and Caucasus in origin, which means that those ancestral groups might have started to expand (numerically) and diversify from there.
Our test is based on the most recent and comprehensive scientific research describing the topology of H haplogroup by Achilli et al. 2004 as well as a few from Loogväli et al. 2004. Upon ordering your test, your DNA will be tested for the following complete list of SNPs that are diagnostic for sub-Haplogroups H1-H15: 7028, 3010, 4769, 951, 750, 6776, 14365, 4336, 3915, 6869, 4793, 13101, 3591, 14470, 13759, 3936, 2259, 11377, 6253.
Your final haplogroup designation will be made by the combined evaluation of the results of these positions and the HVR-I and II information that cover any position between 16001-16569 and 00073-00577 respectively.
An abstract of these papers can be found here.
What does my subclade mean to me? We have drawn together descriptions of each branch of haplogroup H, which we include below. Look for your branch in this chart to read a brief summary of what information is available about it so far. If your specific subclade has only a very brief description, it wasn’t detected enough to offer geographically proximity. This is a failure of the sample size studied and not of your sub-branch. Now that several papers have been published certainly others will follow in short order and we expect that those future papers will offer greater sample size and tighter breakdowns to specific geographical locations.
The following is a simplified chart which displays the positions we currently test and their corresponding haplogroups: :
HAPLOGROUP H DESCRIPTIONS
H - Mitochondrial haplogroup H is a predominantly European haplogroup that participated in a population expansion beginning approximately 20,000 years ago.Today, about 40% of all mitochondrial lineages in Europe are classified as haplogroup H. It is rather uniformly distributed throughout Europe suggesting a major role in the peopling of Europe, and descendant lineages of the original haplogroup H appear in the Near East as a result of migration. Future work will better resolve the distribution and historical characteristics of this haplogroup.
H* - A Haplogroup assignment of H* indicates that you belong to Haplogroup H, but not to any of the subclades you were tested for and that were known at the time that the test was performed. Since new subclades will continue to be discovered, it is unreasonable and expensive to test for each of the additional subclades after each publication. The best way to resolve a sample that is haplogroup H* is through testing the full mtDNA sequence. This test would provide us with all of the mutations in a person’s mtDNA, which means that that person would never need to do any additional mtDNA testing. Even if they are still H* after the full sequence test, their results can be immediately used to attempt to identify a subclade when new subclades are published.
H1 – H1 is the most common branch of haplogroup H. It represents 30% of people in haplogroup H, and 46% of the maternal lineages in Iberia. 13-14% of all Europeans belong to this branch, and H1 is about 13,000 years old.
H1a – H1a is a branch of H1. Further research will better resolve the distribution and historical characteristics of this haplogroup.
H1b – H1b is detected at its highest frequency in Eastern Europe and North Central Europe. It is also found in about 5% of haplogroup H lineages in Siberian Mansis.
H2 – H2 is somewhat common in Eastern Europe and the Caucasus, but likely spread from Western Europe because it is not found in significant frequency in the Near East. It is found in its highest frequency in Germany and Scotland.
H2a – Haplogroup H2a is found most frequently in Eastern Europe, and at a low frequency in Western Europe. Unlike its parent branch H2, H2a’s geographical distribution extends to Central Asia.
H2b – H2b is the branch to which the CRS belongs. Further research will better resolve the distribution and historical characteristics of this haplogroup.
H3 – H3 is the second most common branch of H. Like H1, it is found mainly in Western Europe. However, H3 is not found in significant frequencies in the Near East. It is at its highest frequency in Iberia and Sardinia, and is about 10,000 years old.
H4 – H4 is an uncommon branch and is found at low frequencies in both Europe and the Near East. Further research will better resolve the distribution and historical characteristics of this haplogroup.
H5 – H5 is distributed across Iberia, Central, Eastern, and Southeastern Europe, and is also found at low frequencies in the Near East, where it may have originated.
H5a – H5a is found at its highest frequency in Central Europe and is about 7-8 thousand years old. It is found at low frequency in Europe, and since it is not found or is rare in the Caucasus and the Near East it likely has a European origin.
H6 – H6 is an older branch of haplogroup H. Its age is estimated at around 40,000 years. Studies suggest that this haplogroup is Middle Eastern or Central Asian in origin. It is also found at very low frequencies in Europe. Further research will better resolve the distribution and historical characteristics of this haplogroup.
H6a – H6a has similar distribution to its parent branch H6. Further research will better resolve the distribution and historical characteristics of this haplogroup.
H6c – H6c is found at very low frequency, and can be found in European populations. Further research will better resolve the distribution and historical characteristics of this haplogroup.
H7 – H7 is an uncommon branch and is found at low frequencies in both Europe and the Near East. Further research will better resolve the distribution and historical characteristics of this haplogroup.
H8 – Like H6, H8 has roots in the Near East and Central Asia. It is very uncommon in Europe. Further research will better resolve the distribution and historical characteristics of this haplogroup.
H9 – H9 is an uncommon branch of H. Further research will better resolve the distribution and historical characteristics of this haplogroup.
H10 – H10 is an uncommon branch of H. Further research will better resolve the distribution and historical characteristics of this haplogroup.
H11 – H11 is an uncommon branch of H. Further research will better resolve the distribution and historical characteristics of this haplogroup.
H12 – H12 is an uncommon branch of H. Further research will better resolve the distribution and historical characteristics of this haplogroup.
H13 – H13 is an uncommon branch and is found at low frequencies in Europe, the Near East, and the Caucasus. Further research will better resolve the distribution and historical characteristics of this haplogroup.
H14 – H14 is an uncommon branch of H. Further research will better resolve the distribution and historical characteristics of this haplogroup.
H15 – H15 is an uncommon branch of H. Further research will better resolve the distribution and historical characteristics of this haplogroup.



